![]() If you have chronic insomnia, but no octher psychiatric illnesses, will melatonin help you? This is the question addressed by a recent multi-center study by Geert Mayer and co-workers.
If you have chronic insomnia, but no octher psychiatric illnesses, will melatonin help you? This is the question addressed by a recent multi-center study by Geert Mayer and co-workers.
Chronic insomnia affects a large chunk of the population; papers cited by the authors have found a prevalence around 1 in 3 adults. It is a notoriously persistent condition and treatment is difficult. Many sleeping pills have unacceptable side effects (such as daytime drowsiness) and/or can only be used for short times. Behavioural interventions are difficult to sustain in the long run.
Hence the great interest in understanding how sleep is regulated, so that we can develop medications targeting the sleep cycle itself. Our circadian rhythms, i.e. the rhythms that tell us when it is morning, midday and night, are regulated by a brain area called the suprachiasmatic nucleus, where there is a collection of “clock cells”, that keep time in cycles of approximately 24 h. Another important component is melatonin secretion from the pineal gland, which synchronises the clock to the light/dark cycle of our environment. When it gets dark in the evening, melatonin secretion goes up.
Melatonin. Image from Wikipedia.
So, why not give some extra melatonin 30 minutes before bedtime and see what happens to the sleep patterns? This is what Mayer et al did, with a placebo control group getting an identical-looking pill. Both groups kept doing it for 6 months, and the researchers recorded sleep patterns regularly as well as the subjective experiences of the patients, assessed by a questionnaire. Towards the end, they switched the treatment group over to placebo to see if there were any withdrawal symptoms. The drug they used was not melatonin itself but a synthetic compound called Ramelteon which binds to the same receptor.
What happened? Nothing, pretty much. The researchers monitored 20 different parameters, and found a statistically and clinically significant difference only in one: the latency to persistent sleep after going to bed. Patients on Ramelteon fell sleep faster. But they did not sleep better or longer. They did not have improved alertness or memory and they did not feel better in any way that the researchers asked for. On the other hand, there were just as many “adverse events” in the treatment and control groups, and no signs of withdrawal symptoms, so Ramelteon treatment seems not to be a risky business.
Now, it is necessary to exercise a bit of caution when interpreting a study that has many endpoints at the same time. The more endpoints you include, the higher is the likelihood that one of them will show a significant result purely out of chance. We should therefore be a little skeptical against the positive finding. Ramelteon seems to be in the league of making sacrifices to the god Hypnos – it doesn’t hurt, it could be a little expensive, and it’s not likely to help.

Hypnos (left) with his twin brother Thanatos (representing death). Painting by J W Waterhouse (1849-1917).
Melatonin could be great for some other uses that weren’t tested in this study – against jetlag, perhaps, or if you are having a manic episode and can’t sleep for that reason. The blogosphere’s number one chronobiologist Coturnix has an excellent overview post on sleep, including a bit on melatonin and its potential therapeutic uses.
Full reference:
Mayer G, Wang-Weigand S, Roth-Schechter B, Lehmann R, Staner C, & Partinen M (2009). Efficacy and safety of 6-month nightly ramelteon administration in adults with chronic primary insomnia. Sleep, 32 (3), 351-60 PMID: 19294955

 Posted by evolvingideas
Posted by evolvingideas 

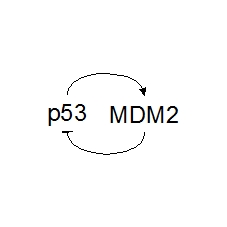 It looks quite straightforward: p53 induces MDM2, which in turn inhibits p53. But can you predict how this system will behave?
It looks quite straightforward: p53 induces MDM2, which in turn inhibits p53. But can you predict how this system will behave?